Dr. Khera is a physician-scientist with expertise in epidemiology, clinical medicine, and human genetics. Among his scientific contributions, he pioneered a new approach to quantify genetic risk for common diseases, […]
Amit V. Khera


Dr. Khera is a physician-scientist with expertise in epidemiology, clinical medicine, and human genetics. Among his scientific contributions, he pioneered a new approach to quantify genetic risk for common diseases, […]
Nat Biotechnol. 2019 Feb 11. doi: 10.1038/s41587-018-0011-0.
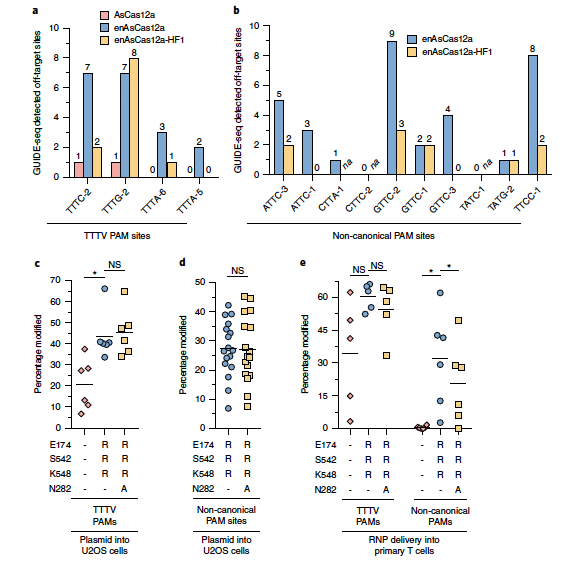
Broad use of CRISPR-Cas12a (formerly Cpf1) nucleases1 has been hindered by the requirement for an extended TTTV protospacer adjacent motif (PAM)2. To address this limitation, we engineered an enhanced Acidaminococcus sp. Cas12a variant (enAsCas12a) that has a substantially expanded targeting range, enabling targeting of many previously inaccessible PAMs. On average, enAsCas12a exhibits a twofold higher genome editing activity on sites with canonical TTTV PAMs compared to wild-type AsCas12a, and we successfully grafted a subset of mutations from enAsCas12a onto other previously described AsCas12a variants3 to enhance their activities. enAsCas12a improves the efficiency of multiplex gene editing, endogenous gene activation and C-to-T base editing, and we engineered a high-fidelity version of enAsCas12a (enAsCas12a-HF1) to reduce off-target effects. Both enAsCas12a and enAsCas12a-HF1 function in HEK293T and primary human T cells when delivered as ribonucleoprotein (RNP) complexes. Collectively, enAsCas12a provides an optimized version of Cas12a that should enable wider application of Cas12a enzymes for gene and epigenetic editing.
NatBiotech-2019-Kleinstiver.pdf
Ben Kleinstiver is a biochemist and genome editor whose interests include translating technologies into molecular medicines. He received his Ph.D in Biochemistry from the University of Western Ontario, and completed […]
Nat Genet. 2017 May 26;49(6):969. PMID: 28546579.
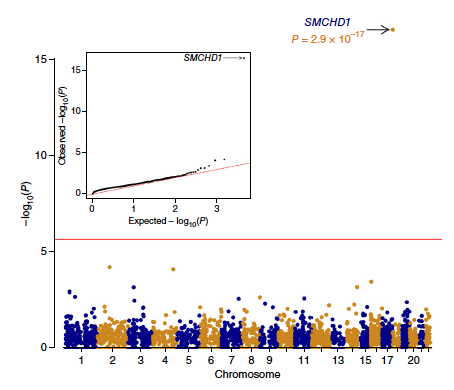
Arhinia, or absence of the nose, is a rare malformation of unknown etiology that is often accompanied by ocular and reproductive defects. Sequencing of 40 people with arhinia revealed that 84% of probands harbor a missense mutation localized to a constrained region of SMCHD1 encompassing the ATPase domain. SMCHD1 mutations cause facioscapulohumeral muscular dystrophy type 2 (FSHD2) via a trans-acting loss-offunction epigenetic mechanism. We discovered shared mutations and comparable DNA hypomethylation patterning between these distinct disorders. CRISPR/Cas9-mediated alteration of smchd1 in zebrafish yielded arhinia-relevant phenotypes. Transcriptome and protein analyses in arhinia probands and controls showed no differences in SMCHD1 mRNA or protein abundance but revealed regulatory changes in genes and pathways associated with craniofacial patterning. Mutations in SMCHD1 thus contribute to distinct phenotypic spectra, from craniofacial malformation and reproductive disorders to muscular dystrophy, which we speculate to be consistent with oligogenic mechanisms resulting in pleiotropic outcomes.
Nat Neurosci. 2017 May;20(5):648-660. doi: 10.1038/nn.4532. Epub 2017 Mar 20.
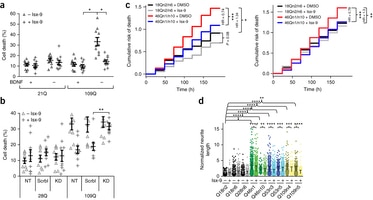
Neural cultures derived from Huntington’s disease (HD) patient-derived induced pluripotent stem cells were used for ‘omics’ analyses to identify mechanisms underlying neurodegeneration. RNA-seq analysis identified genes in glutamate and GABA signaling, axonal guidance and calcium influx whose expression was decreased in HD cultures. One-third of gene changes were in pathways regulating neuronal development and maturation. When mapped to stages of mouse striatal development, the profiles aligned with earlier embryonic stages of neuronal differentiation. We observed a strong correlation between HD-related histone marks, gene expression and unique peak profiles associated with dysregulated genes, suggesting a coordinated epigenetic program. Treatment with isoxazole-9, which targets key dysregulated pathways, led to amelioration of expanded polyglutamine repeat-associated phenotypes in neural cells and of cognitive impairment and synaptic pathology in HD model R6/2 mice. These data suggest that mutant huntingtin impairs neurodevelopmental pathways that could disrupt synaptic homeostasis and increase vulnerability to the pathologic consequence of expanded polyglutamine repeats over time.
Developmental-alterations-in-Huntingtons-disease-neural-cells-and-pharmacological-rescue-in-cells-and-mice.pdf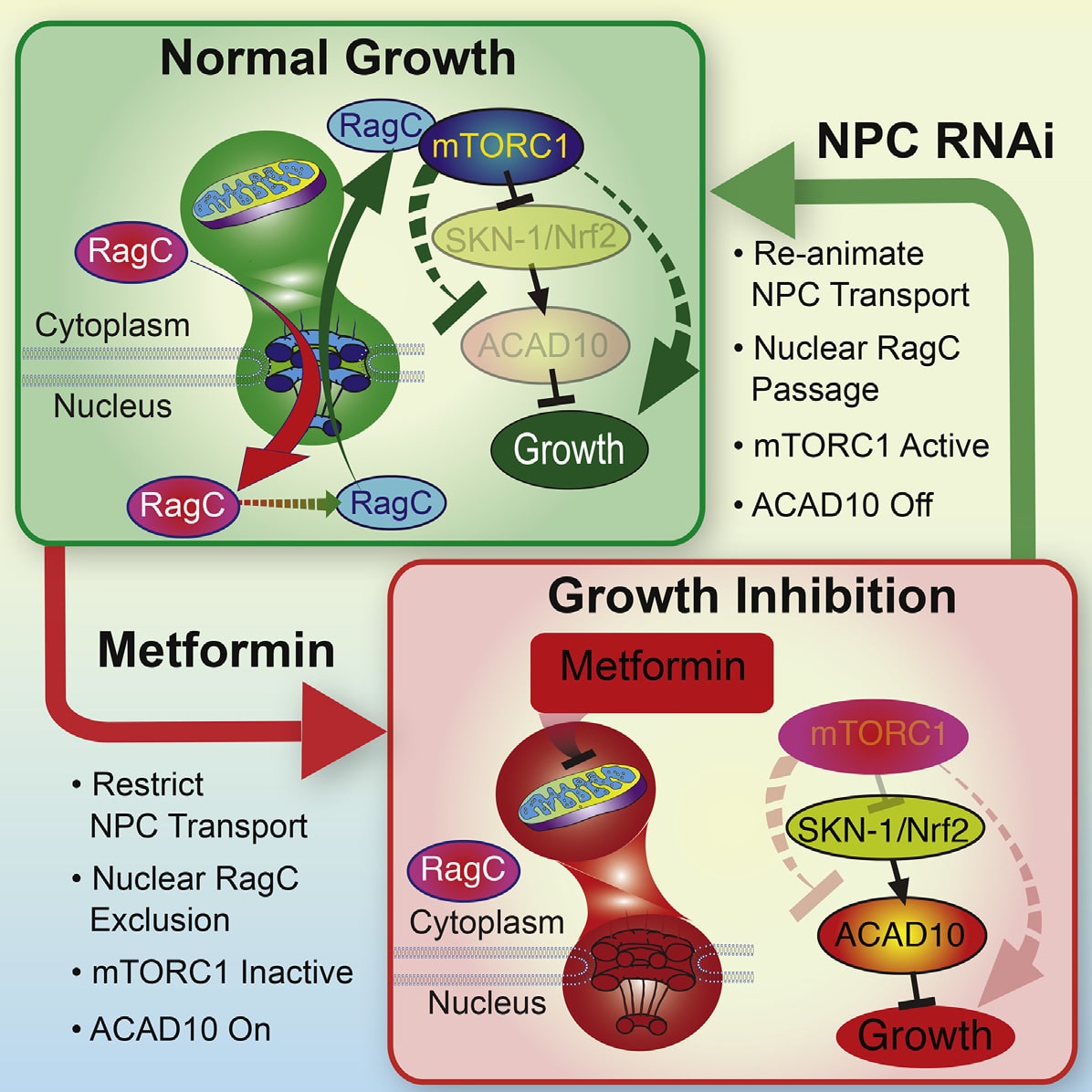
Cell. 2016 Dec; 167(7), p1705–1718.e13.
Metformin has utility in cancer prevention and treatment, though the mechanisms for these effects remain elusive. Through genetic screening in C. elegans , we uncover two metformin response elements: the nuclear pore complex (NPC) and acyl-CoA dehydrogenase family member-10 (ACAD10). We demonstrate that biguanides inhibit growth by inhibiting mitochondrial respiratory capacity, which restrains transit of the RagA-RagC GTPase heterodimer through the NPC. Nuclear exclusion renders RagC incapable of gaining the GDP-bound state necessary to stimulate mTORC1. Biguanideinduced inactivation of mTORC1 subsequently inhibits growth through transcriptional induction of ACAD10. This ancient metformin response pathway is conserved from worms to humans. Both restricted nuclear pore transit and upregulation of ACAD10 are required for biguanides to reduce viability in melanoma and pancreatic cancer cells, and to extend C. elegans lifespan. This pathway provides a unified mechanism by which metformin kills cancer cells and extends lifespan, and illuminates potential cancer targets.
An-Ancient-Unified-Mechanism-for-Metformin-Growth-Inhibition.pdf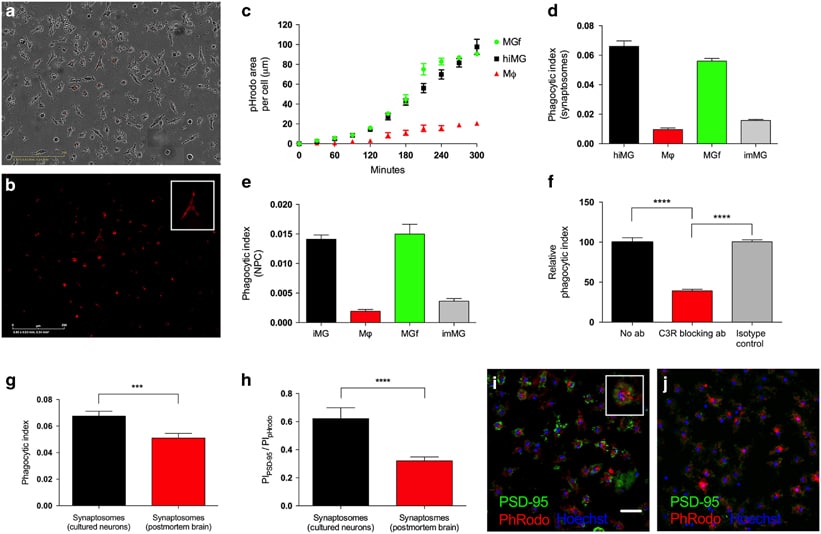
Mol Psychiatry. 2016 Dec 13. doi: 10.1038/mp.2016.220. [Epub ahead of print]
Engulfment of synapses and neural progenitor cells (NPCs) by microglia is critical for the development and maintenance of proper brain circuitry, and has been implicated in neurodevelopmental as well as neurodegenerative disease etiology. We have developed and validated models of these mechanisms by reprogramming microglia-like cells from peripheral blood mononuclear cells, and combining them with NPCs and neurons derived from induced pluripotent stem cells to create patient-specific cellular models of complement-dependent synaptic pruning and elimination of NPCs. The resulting microglia-like cells express appropriate markers and function as primary human microglia, while patient-matched macrophages differ markedly. As a demonstration of disease-relevant application, we studied the role of C4, recently implicated in schizophrenia, in engulfment of synaptic structures by human microglia. The ability to create complete patient-specific cellular models of critical microglial functions utilizing samples taken during a single clinical visit will extend the ability to model central nervous system disease while facilitating high-throughput screening.
Patient-specific-models-of-microglia-mediated-engulfment-of-synapses-.pdf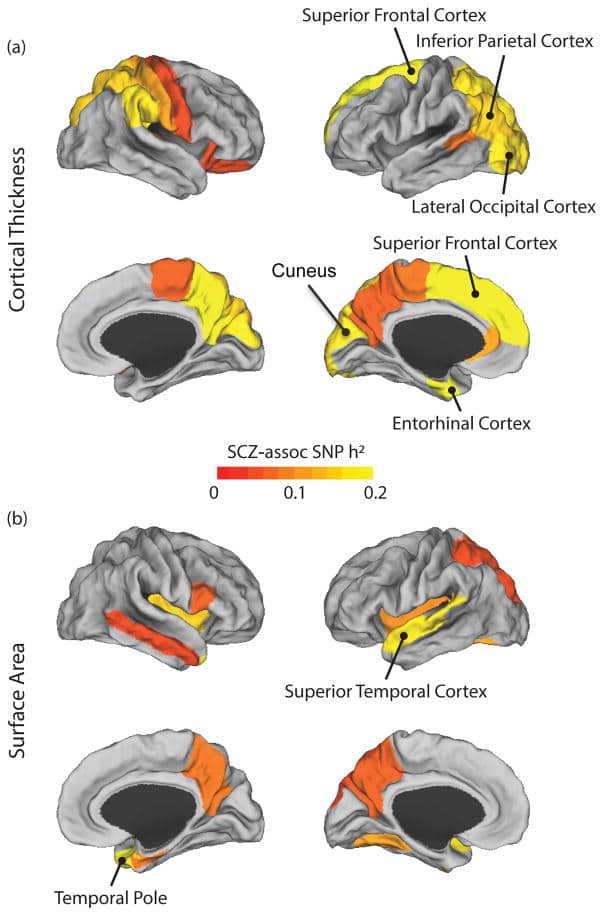
Mol Psychiatry. 2016 Dec;21(12):1680-1689. doi: 10.1038/mp.2016.164. Epub 2016 Oct 11.
Schizophrenia is a devastating neurodevelopmental disorder with a complex genetic etiology. Widespread cortical gray matter loss has been observed in patients and prodromal samples. However, it remains unresolved whether schizophrenia-associated cortical structure variations arise due to disease etiology or secondary to the illness. Here we address this question using a partitioning-based heritability analysis of genome-wide single-nucleotide polymorphism (SNP) and neuroimaging data from 1750 healthy individuals. We find that schizophrenia-associated genetic variants explain a significantly enriched proportion of trait heritability in eight brain phenotypes (false discovery rate=10%). In particular, intracranial volume and left superior frontal gyrus thickness exhibit significant and robust associations with schizophrenia genetic risk under varying SNP selection conditions. Cross-disorder comparison suggests that the neurogenetic architecture of schizophrenia-associated brain regions is, at least in part, shared with other psychiatric disorders. Our study highlights key neuroanatomical correlates of schizophrenia genetic risk in the general population. These may provide fundamental insights into the complex pathophysiology of the illness, and a potential link to neurocognitive deficits shaping the disorder.
Partitioning-heritability-analysis-reveals-a-shared-genetic-basis.pdf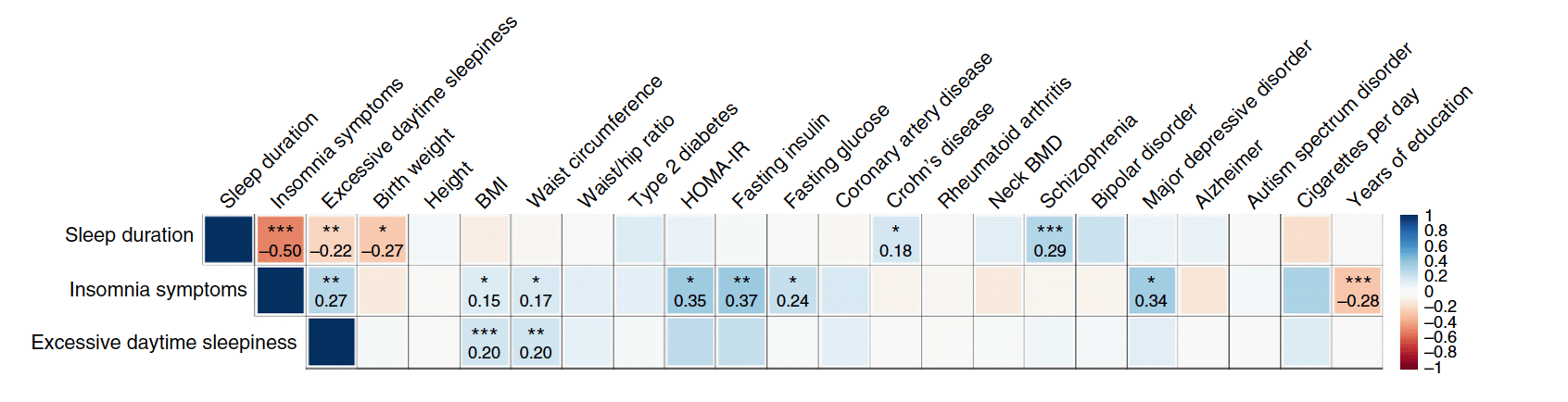
Nat Genet. 2016 Dec 19. doi: 10.1038/ng.3749.
Chronic sleep disturbances, associated with cardiometabolic diseases, psychiatric disorders and all-cause mortality, affect 25-30% of adults worldwide. Although environmental factors contribute substantially to self-reported habitual sleep duration and disruption, these traits are heritable and identification of the genes involved should improve understanding of sleep, mechanisms linking sleep to disease and development of new therapies. We report single- and multiple-trait genome-wide association analyses of self-reported sleep duration, insomnia symptoms and excessive daytime sleepiness in the UK Biobank (n = 112,586). We discover loci associated with insomnia symptoms (near MEIS1, TMEM132E, CYCL1 and TGFBI in females and WDR27 in males), excessive daytime sleepiness (near AR-OPHN1) and a composite sleep trait (near PATJ (INADL) and HCRTR2) and replicate a locus associated with sleep duration (at PAX8). We also observe genetic correlation between longer sleep duration and schizophrenia risk (rg = 0.29, P = 1.90 × 10-13) and between increased levels of excessive daytime sleepiness and increased measures for adiposity traits (body mass index (BMI): rg = 0.20, P = 3.12 × 10-9; waist circumference: rg = 0.20, P = 2.12 × 10-7).
Lane_sleep_disturbance_GWAS_2016.pdf